Stanford’s Mahajan Lab’s new artificial intelligence (AI) driven eye “aging clock” has revealed that diseased eyes undergo accelerated aging, particularly within disease-specific cell types.
The study was published in Cell in October and featured at the National Institutes of Health (NIH) Tuesday. The team integrated proteomics techniques to create a software-based approach called tracing expression of multiple protein origins (TEMPO), capable of analyzing protein levels in only a few drops of eye fluid from a liquid biopsy.
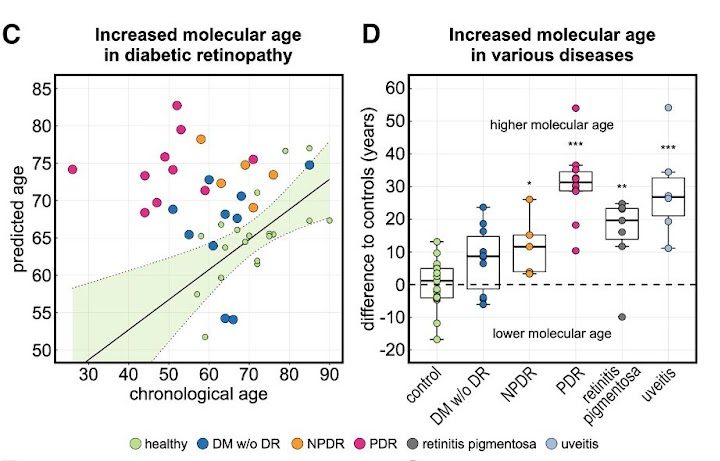
This analysis mapped almost 6,000 proteins to their cellular origins, allowing for the construction of a machine learning model that could predict a patient’s age based on the expression levels of just 26 of these proteins. The algorithm predicted the molecular age of the eye to be at times decades older in diseased patients compared to healthy ones.
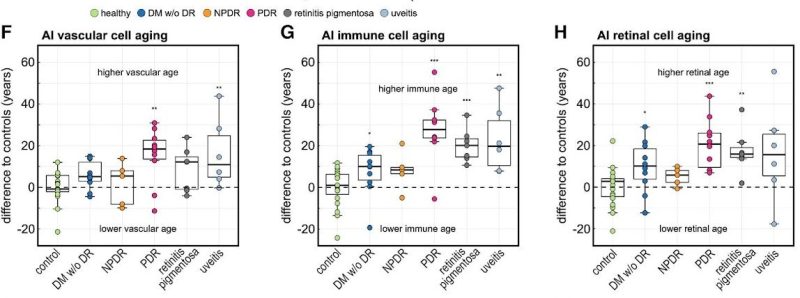
The researchers also found that eye diseases were associated with accelerated aging of disease-specific cell types.
Vinit Mahajan, an ophthalmology professor and an author of the study, said he was inspired to develop this “aging clock” when he was invited to speak at a longevity meeting. There, he learned about other Stanford researchers within the field also developing aging clocks.
To make the clock a reality, the lab collaborated with professors and graduate students from the biomedical, data science and computer science departments to develop the final AI model.
“Stanford is such a collegial campus. We really worked across departments,” Mahajan said.
The lab obtained 120 eye fluid samples from patients undergoing eye surgery.
From each patient’s liquid biopsy, the team detected and measured the levels of approximately 6,000 different proteins using a DNA aptamer-based assay. The team employed single-cell RNA sequencing to trace each protein to its cellular origin.
“One of the reasons why we could detect aging patterns was a question of proteomic resolution,” said Julian Wolf, an ophthalmology postdoctoral fellow and the primary author of the study.
The TEMPO software allowed for a high resolution, minimally invasive look inside living cells using only 50 microliters of eye fluid, eliminating the need for a potentially damaging tissue biopsy of the eye or postmortem tissue analysis.
The TEMPO approach shows promise for enhancing patient selection in clinical trials, especially in cases where drugs target proteins whose levels might not necessarily be different between diseased and healthy patients.
“We think it is very valuable to look at the molecular level to determine if the protein targets of drug treatments are actually there,” Wolf said. “We could have a better tool to decide which drug is good for which patient.”
The lab incorporated artificial intelligence to predict the age of the eye based on protein biomarkers from the liquid biopsies. The team divided all 120 liquid biopsy samples into healthy and diseased cohorts. They trained and validated the model on the healthy samples.
A cross-validation AI model separated all healthy samples into 10 equal subgroups, or folds. For nine folds, the model was fed the sample’s protein expression levels, quantified with fluorescence intensity values, and then provided the patient’s birthday. The model was validated on the 10th fold, where it predicted the patient’s age on the input proteomics data alone.
Using feature-selection methods, the algorithm whittled down the 6,000 proteins to the 26 most important ones, whose given levels allowed the algorithm to accurately predict that patient’s age. Of the 26 proteins, 20 were found to have known associations with aging.
The team then applied the AI model on patients with eye-diseases not primarily related to age, such as diabetic retinopathy, retinitis pigmentosa and uveitis. They found that the molecular age predicted by the model was at times decades older than the actual chronological age of the patient, even among patients who had already responded positively to clinical treatments. Mahajan said this finding suggests the need for “supplemental anti-aging therapies” to ensure a patient’s full recovery.
In patients with late-stage, proliferative diabetic retinopathy — an eye condition that causes vision loss in diabetic patients — the algorithm predicted the molecular age of the eye to be about 30 years older than those patients’ chronological ages. For example, if a patient’s actual age was 50, the algorithm predicted their age to be 80, because they were expressing proteins in the same way that an 80-year-old would, Mahajan said.
“This is concrete molecular data that says yes, disease accelerates aging,” Mahajan said.
Among patients with Parkinson’s disease, the study also found that disease-linked brain proteins were detectable in the eye fluid samples. This finding suggested that protein biomarkers in the eye could serve as a window to many other conditions.
The team’s next goal is to increase the sample size to refine their model and apply the TEMPO approach to bodily fluids and diseases outside of the eye.
“In principle, the TEMPO approach can be applicable to completely unrelated organ systems,” Wolf said. “For example, collecting urine might tell you something about the kidney, or collecting spinal fluid might tell you something about the brain.”
Jeffrey Goldberg, professor and chair of ophthalmology at the Byers Eye Institute, who was unaffiliated with the study, emphasized the novelty of its approach to medicine.
“This work describes a paradigm shift in how we study humans and disease and then apply this new approach to uncover pathways at the molecular level important in aging. The broad future dissemination of [the] work will change how we look at any disease process,” Goldberg said.
